Individual Ion Mass Spectrometry
Description: Charge detection mass spectrometry (CDMS) has enabled the generation of true mass spectra with the direct readout of an ion’s integer charge value (for example 35+, 34+, and so on). We have shown that the commercially available Orbitrap mass analyzer can multiplex and regularize CDMS for creation of mass spectra on extremely complex samples. Individual ion analysis not only determines the integer charge value for each ion, but results in enhanced resolution and 500x dilute sample compatibility. This I2MS approach is shown to be general for measuring complex proteoform mixtures and their complexes, covering a range of masses from 8 kDa to 3.2 MDa. Orbitrap-based I2MS is capable of precise mass determination where ensemble measurements fail or require prior separation. I2MS analysis is highly applicable to complex denatured proteoform mixtures, VLP engineering, can be extended to other promising systems including Qβ virions, adeno-associated virus (AAV) and hepatitis B, and can yield important information for more complex systems, including bacterial microcompartments that contain an assortment of coat-protein sizes.
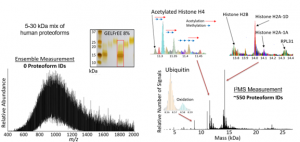
I2MS analysis mass output (right) reveals hundreds of proteoforms from the complex landscape of a 5-30 kDa mix of HEK-293 cell lysate. Traditional ensemble mass spectrometry measurement in the m/z domain (left) is too complex for any analogous proteoform assignment yielding 0 identifications.
TR&D 7 Development Status: On going
Selected Publications (3/7):
1.Kafader, J. O; Melani, R. D; Durbin, K. R; Ikwuagwu, B; Early, B. P; Fellers, R. T; Beu, S. C; Zabrouskov, V; Makarov, A. A; Maze, J. T; Shinholt, D. L; Yip, P. F; Tullman-Ercek, D; Senko, M. W; Compton, P. D; Kelleher, N. L., Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes. Nat Methods., 17 (4), pp. 391-394, 2020.
2.Kafader, J. O; Durbin, K. R; Melani, R. D; Des Soye, B. J; Schachner, L. F; Senko, M. W; Compton, P. D; Kelleher, N. L., Individual Ion Mass Spectrometry Enhances the Sensitivity and Sequence Coverage of Top-Down Mass Spectrometry. J Proteome Res., 19 (3), pp. 1346-1350, 2020.
3.McGee, J. P; Melani, R. D; Yip, P. F; Senko, M. W; Compton, P. D; Kafader, J. O; Kelleher, N. L., Isotopic Resolution of Protein Complexes up to 466 kDa Using Individual Ion Mass Spectrometry. Anal Chem., 93 (5), pp. 2723-2727, 2021.
Scientific Presentations:
ASMS 2021, ASMS 2020, World HUPO 2019, ASMS 2019



Comments are closed.