Top and Middle Down of p53 Modification Codes
In collaboration with:
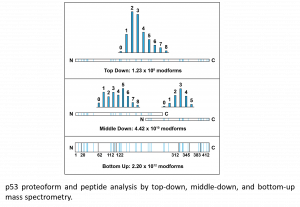
Description: Post-translational modifications (PTMs) are found on most proteins, particularly on “hub” proteins like the tumor suppressor protein, p53. p53 has >100 potential PTM sites and substantial crosstalk among PTM sites somehow underlies its ability to integrate diverse signals and coordinate effector functions. Disentangling the combinatorial explosion in PTM patterns (“modforms”) across an entire protein is challenging, as peptide-based mass spectrometry strategies (bottom-up proteomics) degrades information on PTM stoichiometry and combinations of PTMs. Alternatively, direct analysis of intact proteins using top-down mass spectrometry captures PTM combinations and stoichiometry, but with limited dynamic range. A combination of three approaches, top-down, middle-down, and bottom-up proteomics, will enable the investigation into p53 PTMs.
Project status: On going
Publications:
-Estimating the Distribution of Protein Post-Translational Modification States by Mass Spectrometry. Philip D. Compton, Neil L. Kelleher, Jeremy Gunawardena
-How many human proteoforms are there? Ruedi Aebersold et al.
Congress Presentations:
– ASMS 2020
Spinoff funding: R01





Comments are closed.