Targeted Top Down Proteomics
Overview
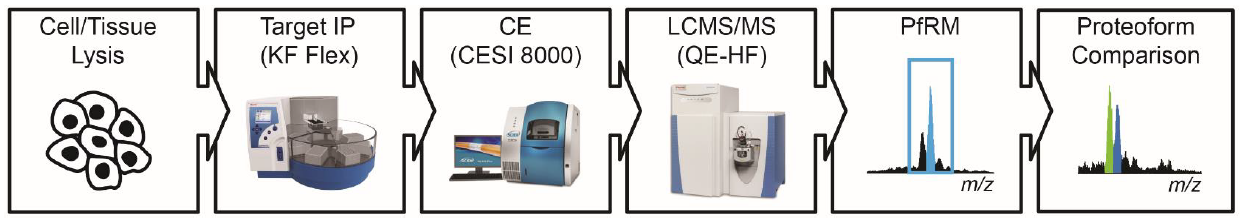
The NRTDP has invested significant resources into a discovery pipeline that efficiently generates interesting candidate proteoforms for further investigation. While these experiments produce valuable insight into cell and disease biology, they are labor intensive and relatively low throughput, thus rendering application of this pipeline to larger sample cohorts significantly challenging. Therefore, we propose the development of a complementary targeted quantitative workflow enabling further interrogation of interesting proteoforms in a high throughput modality with enhanced sensitivity. Automated immunoprecipitation will provide the foundation of this new platform. Here, we will draw on the experience of our collaborators at Thermo Pierce, who have validated thousands of antibodies and implemented automated immunocapture using the KingFisher Flex system. Once immunoprecipitated, samples will be resolved via liquid chromatography, ultimately transitioning to the CESI 8000 capillary electrophoresis system from Sciex for improved sensitivity. For each target, we will develop a proteoform reaction monitoring (PfRM) assay that validates and quantitates all of the proteoforms present at stoichiometric levels. The resulting data will be processed using a new software platform, PFRM VIEWER, with the finalized PfRM assays made freely available to the scientific community through a portal on the Resource website. These combined innovations will enable the Resource to advance further down the pathway to translational proteomics by confirming observations in larger populations while also providing a roadmap for community members to implement these assays within their own respective research initiatives.
Capillary Electrophoresis
Immunoprecipitated proteoforms will be concentrated and desalted by C4 ZipTip, resolved by CE using a CESI 8000, and introduced into a QE-HF mass spectrometer for analysis by targeted TDMS. We anticipate that each target proteoform population will exhibit distinct properties, thus necessitating CE parameter optimization for each target rather than analysis by a universally applicable method. We will therefore employ recombinant versions of target proteoforms such as p53 (DBP 2) or ApoC-III (DBP 14), either unmodified or modified in vitro, to determine which column coatings, loading methods, and separation conditions provide optimal loading and resolution of these species. The latter parameter will be most significant for proteoform mixtures, as each component should elute within a distinct migration time window for optimal application of our targeted MS approach.


Comments are closed.