Characterization of RAS Family Proteins in Colorectal Cancer
In collaboration with:
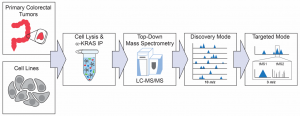
Description: The characterization of biologically relevant post-translational modifications (PTMs) on RAS has historically been carried out through methodologies such as immunoblotting with PTM-specific antibodies or peptide-based proteomic methods. While these methods have the potential to identify a given PTM on KRAS4B, they are incapable of characterizing or distinguishing the different molecular forms or proteoforms of KRAS4B from those of related RAS isoforms. We have further optimized our IP-TDMS protocol to enable us to characterize KRAS4A, KRAS4B, and HRAS proteoforms, including those present at <5% relative abundance. Most recently, we deployed this optimized RAS proteoform assay to a panel of 12 cell lines and 34 colorectal tumor samples in a collaboration with the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and NCI RAS Initiative.
Experimental workflow schematic for RAS IP-TDMS assays which couples immunoprecipitation of with downstream top-down mass spectrometry.
Project status: Ongoing
Publications:
-Precise Characterization of KRAS4B Proteoforms by Combining Immunoprecipitation with Top-Down Mass Spectrometry. 2021. DOI: 10.1007/978-1-0716-1190-6_3
Congress Presentations:
-Third RAS Initiative Symposium 2021
-FASEB Cell Signaling in Cancer Conference: From Mechanisms to Therapy 2020





Comments are closed.