21st Century Analysis of Histone Modifications
Overview
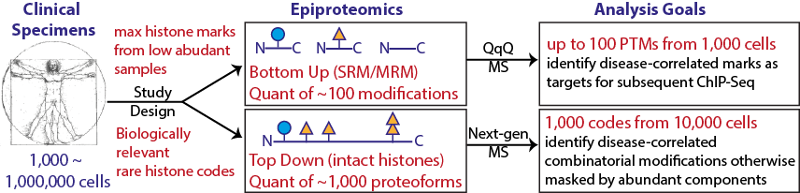
It is well-known that distinct histone modification states carry key information regarding gene transcription. Histone methylation, in particular, can transmit information through mitosis, forming the basis of an epiproteomic “fingerprint.” In TR&D 4, we will establish and validate a full pipeline to quantitate all major sites of lysine methylation from 106 down to 103 cells. The output of this pipeline will be a quantitative report of modification levels spanning 5 orders of magnitude in abundance for >100 discrete modification states. To accomplish this feat, we propose a bifurcated approach using two different mass spectrometric strategies for protein analysis. The platform will include interrogating full-length histones (top-down) and triple-quadrupole based tryptic peptide quantitation (bottom-up).
Top Down Analysis of Histone H3
Histone H3 is the most highly modified core histone protein. In a recent publication in Molecular and Cellular Proteomics, we brought together the collaborating lab of Dr. Jonathan Licht with our instrumentation and data analysis teams to explore the different histone proteoforms present in a multiple myeloma cell line. The result was a deep and thorough analysis of nearly 100 different proteoforms of histone H3.1 and H3.3 where we recapitulated the antagonistic crosstalk between K27 and K36 methylation in H3.1.


Comments are closed.